ABFE¶
This document describes how to run a ABFE simulation using Deep Origin tools.
Prerequisites¶
We assume that we have an initialized and configured Complex object:
from deeporigin.drug_discovery import Complex, BRD_DATA_DIR
sim = Complex.from_dir(BRD_DATA_DIR)
Here, ABFE requires that the Complex object have an already prepared protein (PDB), and the associated ligands (SDF) are in a docked pose.
Warning
The Complex.from_dir() function only accepts 1 PDB file per directory. This function will throw an error if it finds more than 1 PDB file per directory.
For more details on how to get started, see Getting Started .
System Preparation¶
First, make sure you have prepared your system and verified that everything is as expected. To prepare a system for a single ligand, use:
ligand = sim.ligands[0]
prepared_system = sim.prepare(ligand=ligand)
prepared_system.show()
You will see something like:
Estimating costs¶
Before starting a ABFE run, you can estimate costs using:
# assuming we want to perform a single ABFE run on a single ligand
ligand = sim.ligands[0]
jobs = sim.abfe.run(ligand=ligand, quote=True)
job = jobs[0]
You will get back a widget representing this job such as this:
Example widget
Prices shown here are for demonstrative purposes only. Actual prices can vary.
Note that this Job is ready to run, but will not actually run unless you approve the amount and confirm.
Starting an ABFE run¶
Confirming a quoted Job¶
If you have already generated a quoted Job (using quote as shown above), you can start the ABFE run using:
job.confirm()
This will start the ABFE run and the job widget will now display:
Multiple ligands¶
To run an end-to-end ABFE workflow on multiple ligands, we use:
jobs = sim.abfe.run(ligands=[sim.ligands[0],sim.ligands[1]])
Omitting the ligand will run ABFE on all ligands in the Complex object.
jobs = sim.abfe.run()
Each ligand will be run in parallel on a separate instance, and each Job can be monitored and controlled independently.
Watch Jobs¶
To monitor the status of this job, use:
job.watch()
This makes the widget auto-update, and monitor the status of the job till it reaches a terminal state (Cancelled, Succeeded, or Failed).
Monitoring Jobs
For more details about how to monitor jobs, look at this How To section.
Parameters¶
Viewing parameters¶
The end to end ABFE tool has a number of user-accessible parameters. To view all parameters, use:
from deeporigin.drug_discovery import Complex, BRD_DATA_DIR
sim = Complex.from_dir(BRD_DATA_DIR)
sim.abfe._params["end_to_end"]
Expected output
This will print a dictionary of the parameters used for ABFE, similar to:
{
"binding": {
"add_fep_repeats": 0,
"annihilate": true,
"restraints_type": "rigid_body",
"em_all": true,
"em_solvent": true,
"emeq_md_options": {
"T": 298.15,
"cutoff": 0.9,
"dt": 0.004,
"fourier_spacing": 0.12,
"hydrogen_mass": 2.0
},
"lambda_schedule": "default",
"n_windows": 32,
"mbar": 1,
"npt_reduce_restraints_ns": 2.0,
"nvt_heating_ns": 1.0,
"prod_md_options": {
"T": 298.15,
"barostat": "MonteCarloBarostat",
"barostat_exchange_interval": 1150,
"cutoff": 0.9,
"dt": 0.004,
"fourier_spacing": 0.12,
"hydrogen_mass": 2.0,
"integrator": "BAOABIntegrator"
},
"repeats": 1,
"replex_period_ps": 0,
"softcore_alpha": 0.5,
"steps": 1250000,
"test_run": 0,
"thread_pinning": 1
},
"solvation": {
"add_fep_repeats": 0,
"annihilate": true,
"restraints_type": "rigid_body",
"em_all": true,
"em_solvent": true,
"emeq_md_options": {
"T": 298.15,
"cutoff": 0.9,
"dt": 0.004,
"fourier_spacing": 0.12,
"hydrogen_mass": 2.0
},
"lambda_schedule": "default",
"n_windows": 24,
"mbar": 1,
"npt_reduce_restraints_ns": 0.2,
"nvt_heating_ns": 0.1,
"prod_md_options": {
"T": 298.15,
"barostat": "MonteCarloBarostat",
"barostat_exchange_interval": 1150,
"cutoff": 0.9,
"dt": 0.004,
"fourier_spacing": 0.12,
"hydrogen_mass": 2.0,
"integrator": "BAOABIntegrator"
},
"repeats": 1,
"softcore_alpha": 0.5,
"steps": 500000,
"test_run": 0,
"thread_pinning": 1
}
}
Modifying parameters¶
Any of these parameters are modifiable using dictionary access. For example, to change the number of windows in the binding step, we can use:
from deeporigin.drug_discovery import Complex, BRD_DATA_DIR
sim = Complex.from_dir(BRD_DATA_DIR)
sim.abfe._params["end_to_end"]["binding"]["n_windows"] = 24
Changing parameters may lead to simulation failures
Some parameters, like dt are restricted to certain ranges. You will not be allowed to start a simulation run if these parameters exceed those ranges.
Changing parameters away from the defaults may lead to simulation failures.
Results¶
Viewing results¶
After initiating a run, we can view results using:
df = sim.abfe.show_results()
df
This shows a table similar to:
| dG | Std | AnalyticalCorr | Repeats | SMILES | r_exp_dg |
|---|---|---|---|---|---|
| -9.98 | 0.0 | -11.46 | 1 | COCCn1cc(-c2cccc(C(=O)N(C)C)c2)c2cc[nH]c2c1=O | -7.22 |
Viewing trajectories¶
To view MD trajectories from this run, refer to this How-to section
Viewing overlap matrix¶
An FEP overlap matrix is a diagnostic used in free energy perturbation calculations to evaluate how well neighboring λ states sample overlapping regions of configuration space. Each matrix element measures the statistical overlap between configurations from different states based on their energy distributions. The goal is to ensure every state has overlap with its neighbors in both directions – so that off-diagonal elements are sufficiently larger than zero.
The overlap matrix between windows can be viewed using:
sim.abfe.show_overlap_matrix(ligand=ligand, run="binding")
An image such as the following will be shown:
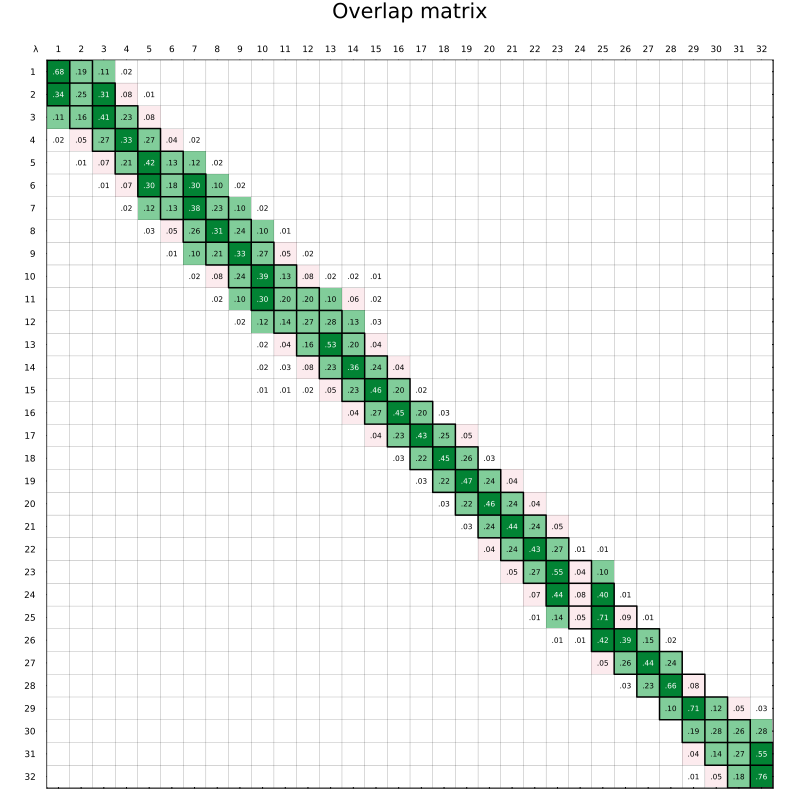
To view the overlap matrix for the solvation run, use:
sim.abfe.show_overlap_matrix(ligand=ligand, run="solvation")
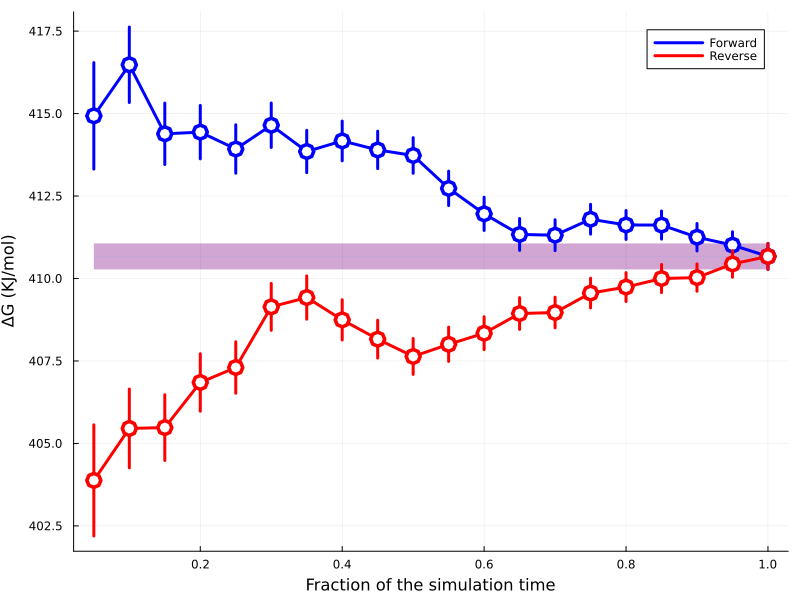
Viewing convergence time plots¶
To view the convergence time plots for a run, use:
sim.abfe.show_convergence_time(ligand=ligand, run="binding")