Visualize proteins and ligands
This document describes how to visualize proteins and ligands constructed using the Protein and Ligand classes, and tools to visualize SDF files.
Visualizing a protein¶
Browser support
These visualizations work best on Google Chrome. We are aware of issues on other browsers, especially Safari on macOS.
A protein object can be visualized using show:
from deeporigin.drug_discovery import Protein
protein = Protein.from_pdb_id("1EBY")
protein.show()
A visualization such as this will be shown:
Jupyter notebook required
Visualizations such as these require this code to be run in a jupyter notebook. We recommend using these instructions to install Jupyter.
Ligands¶
Ligand objects¶
The Ligand class is the primary way to work with ligands in Deep Origin.
Browser support
These visualizations work best on Google Chrome. We are aware of issues on other browsers, especially Safari on macOS.
A ligand object can be visualized using show:
from deeporigin.drug_discovery import Ligand
serotonin = Ligand.from_identifier("serotonin")
serotonin.show()
If a ligand is backed by a SDF file, a 3D visualization will be shown, similar to:
A visualization such as this will be shown:
Jupyter notebook required
Visualizations such as these require this code to be run in a jupyter notebook. We recommend using these instructions to install Jupyter.
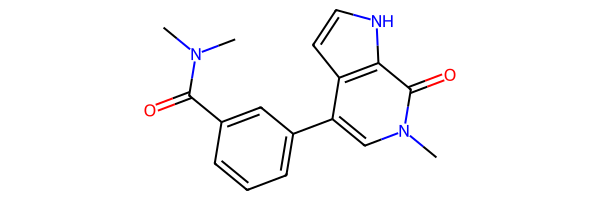
If a ligand is not backed by a SDF file, a 2D visualization will be shown: