ABFE¶
This document describes how to run a ABFE simulation using Deep Origin tools.
Prerequisites¶
Make sure you have installed, configured, and authenticated with the Deep Origin python client.
Background¶
The following flowchart describes the workflow of ABFE that we will go through. Square boxes represent tools in the workflow, and rounded rectangles represent artifacts and files.
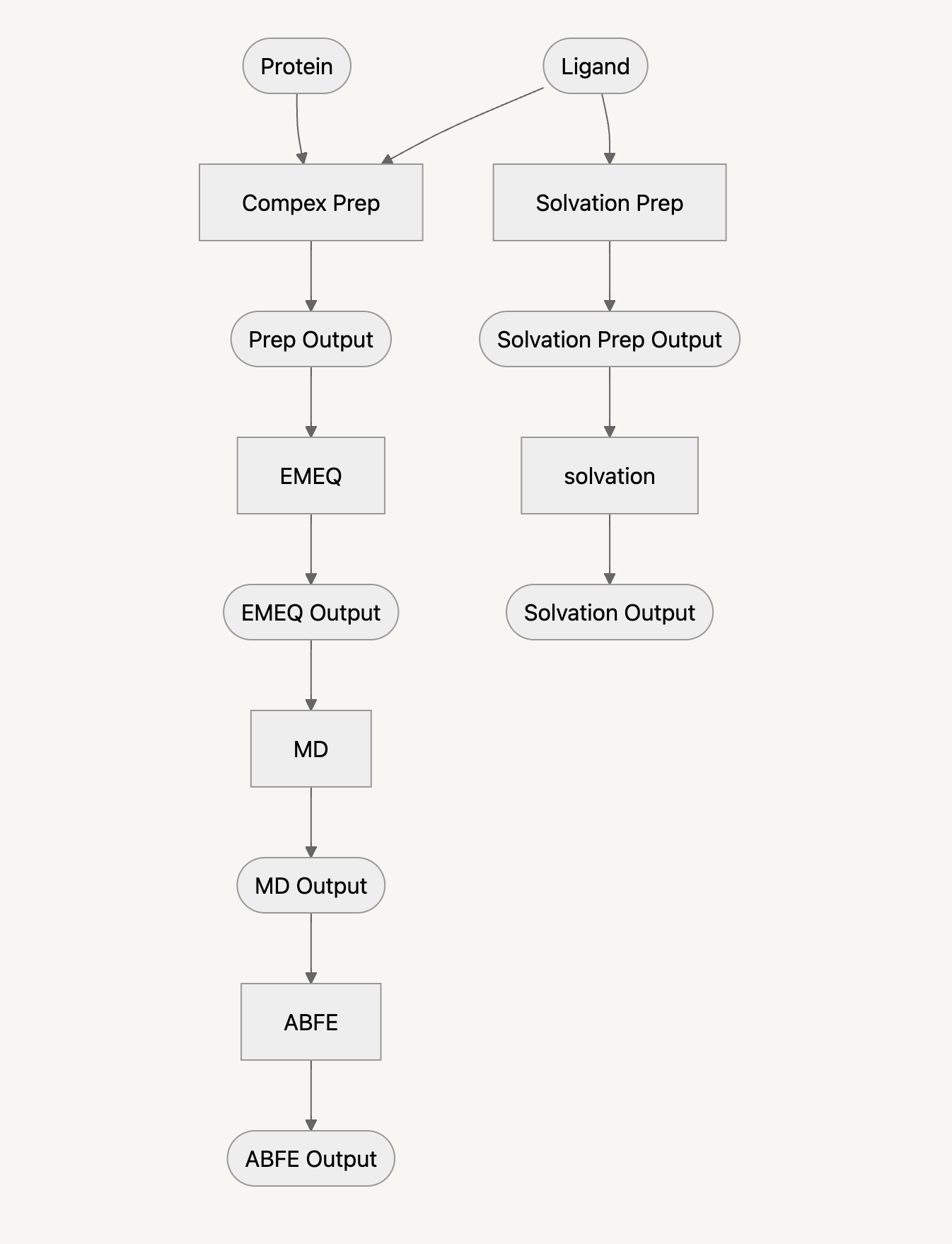
Required input files¶
You will need to have the following input files on your local computer:
- A ligand file, in SDF format
- A protein PDF file
Running ABFE workflow¶
Jupyter notebooks
It is assumed that you are working in a Jupyter notebook (or similar IPython environment). This makes it easier to run the workflow, and some functions, such as wait_for_jobs assume that you are in a Jupyter notebook.
First, we import necessary functions and modules from the deeporigin package:
from deeporigin.tools.utils import wait_for_jobs
from deeporigin.tools import abfe
We then specify where our input files are:
protein_file = "/path/to/protein.pdb"
ligand_file = "/path/to/ligand.sdf"
Initialization¶
Here, we data structures to store input and intermediate files, and upload input files to Deep Origin:
row_id = abfe.init(
ligand_file=ligand_file,
protein_file=protein_file,
)
You will see a message similar to:
Using database at: https://os.deeporigin.io/org/org/data/database/ABFE
🧬 Uploading files to database...
🧬 Files uploaded to row ABFE-1.
This function creates a database for you on the DataHub, generates necessary columns, and uploads your input ligand and protein files to the appropriate columns in that database.
The created database will look something like this:
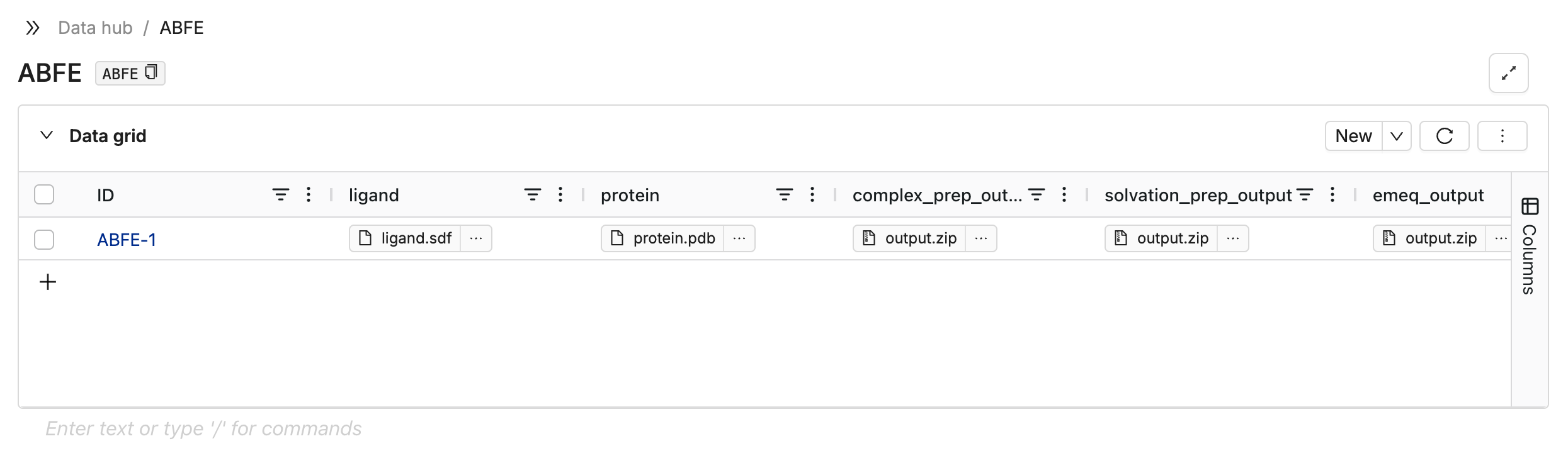
Row IDs
The row_id identifies a row in this database, and is used by all subsequent functions. You will always need to specify the row_id to each function.
Complex Prep¶
To run complex prep on the ligand and protein file, we use:
abfe.complex_prep(
row_id=row_id,
is_lig_protonated=True,
)
This queues up a task on Deep Origin. When it completes, outputs will be written to the appropriate column in this database.
You will see a message printed to screen similar to:
Using row ABFE-1 in database
🧬 Job started with ID: 20f05e96, execution ID: x9rl5eghrpqwyiciehc3e
Parameters
This function exposes a number of parameters. To see which parameters are available, and to modify them, look at the reference documentation
Solvation Prep¶
We do not have to wait for the complex prep step to complete in order to start this step. So we can start this step using:
abfe.solvation_prep(row_id=row_id)
This queues up a task on Deep Origin. When it completes, outputs will be written to the appropriate column in this database.
You will see a message printed to screen similar to:
Using row ABFE-1 in database
🧬 Job started with ID: 20f05e96, execution ID: x9rl5eghrpqwyiciehc3e
Parameters
This function exposes a number of parameters. To see which parameters are available, and to modify them, look at the reference documentation
Waiting for Jobs
At this stage, we need to wait for these steps to complete to before proceeding to the next step. For example, we need to wait for the Complex Prep step to complete in order to proceed to the EMEQ step.
To do this, use the wait_for_jobs function. The wait_for_jobs repeatedly polls Deep Origin to determine the status of jobs, and terminates when all jobs have completed.
While it is running (and blocking execution), you will see a table similar to this:
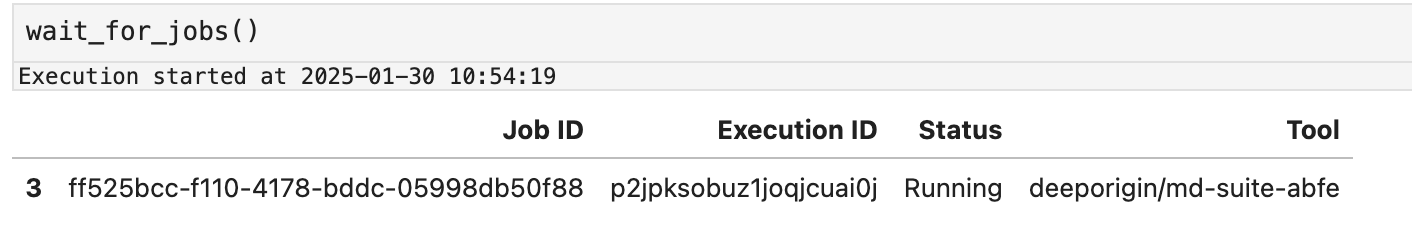
EMEQ¶
To run EMEQ, we use:
abfe.emeq(row_id=row_id)
This queues up a task on Deep Origin. When it completes, outputs will be written to the appropriate column in this database.
You will see a message printed to screen similar to:
Using row ABFE-1 in database
🧬 Job started with ID: 20f05e96, execution ID: x9rl5eghrpqwyiciehc3e
Parameters
This function exposes a number of parameters. To see which parameters are available, and to modify them, look at the reference documentation
Simple MD¶
To run simple MD, we use:
abfe.simple_md(
row_id=row_id,
steps=1000,
)
This queues up a task on Deep Origin. When it completes, outputs will be written to the appropriate column in this database.
You will see a message printed to screen similar to:
Using row ABFE-1 in database
🧬 Job started with ID: 20f05e96, execution ID: x9rl5eghrpqwyiciehc3e
Parameters
This function exposes a number of parameters. To see which parameters are available, and to modify them, look at the reference documentation
Binding FEP¶
To run Binding FEP, we use:
abfe.binding_fep(
row_id,
steps=1000,
)
This queues up a task on Deep Origin. When it completes, outputs will be written to the appropriate column in this database.
You will see a message printed to screen similar to:
Using row ABFE-1 in database
🧬 Job started with ID: 20f05e96, execution ID: x9rl5eghrpqwyiciehc3e
Parameters
This function exposes a number of parameters. To see which parameters are available, and to modify them, look at the reference documentation
Solvation FEP¶
To run Solvation FEP, we use:
abfe.solvation_fep(
row_id=row_id,
annihilate=True,
)
This queues up a task on Deep Origin. When it completes, outputs will be written to the appropriate column in this database.
You will see a message printed to screen similar to:
Using row ABFE-1 in database
🧬 Job started with ID: 20f05e96, execution ID: x9rl5eghrpqwyiciehc3e
Parameters
This function exposes a number of parameters. To see which parameters are available, and to modify them, look at the reference documentation
Inspecting outputs¶
The output of any step can be inspected by downloading the linked zip file in the appropriate column in the database.